Real-time
Quantitative PCR
|
Cat.
No. SER006
|
Service
|
Major
Service Items
-
Detection
System: LightCycler 1.5 (Roche)
-
Primer
& probe design
-
Quantitation
assay (SYBR Green, Universal Probe Library
& hybridization probe system)
-
Melting
curve
-
PCR
efficiency calibration
|
Our
Experience
|
|
Between Run: CV < 2%
Within
Run: CV < 1 %
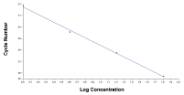
PCR Efficiency = 1.99 (As Figure) |
 |
Applications
-
Gene expression analysis,
such as oncological or virological research
parameters
-
Validation of DNA
microarray result
-
Validation of gene
knock-down result
-
Food pathogens &
spoilage testing
-
Screening for genetically modified organisms (GMO)
|
Background
information
The real-time PCR system is based on the detection and
quantitation of a fluorescent reporter (Lee,
1993; Livak,
1995). Advantages are:
-
Traditional PCR is measured at end-point (plateau),
while real-time PCR collects data in the exponential
growth phase
-
An increase in reporter fluorescent signal is directly
proportional to the number of amplicons generated
-
The cleaved probe provides a permanent record
amplification of an amplicon
-
Requirement of 1000-fold less RNA than conventional
assays
-
No-post PCR processing (no
electrophoretical separation of amplified DNA)
-
Detection is capable down to a 2-fold change.
References
-
Lee et al. (1993) Allelic discrimination by nick-translation PCR with
fluorogenic probes. Nucleic
Acids Res, 21, 3761-3766.
-
Livak et al. (1995)
Oligonucleotides with fluorescent dyes at opposite ends
provide a quenched probe system useful for detecting PCR product
and nucleic acid hybridization. PCR
Methods Appl, 4, 357-362.
|

